http://english.cas.cn/newsroom/research_news/life/202510/t20251010_1089023.shtml
A research team from the Qingdao Institute of Bioenergy and Bioprocess Technology (QIBEBT) of the Chinese Academy of Sciences has identified a specific histone modification as the key regulator governing microalgae’s adaptation to low-CO2environments.
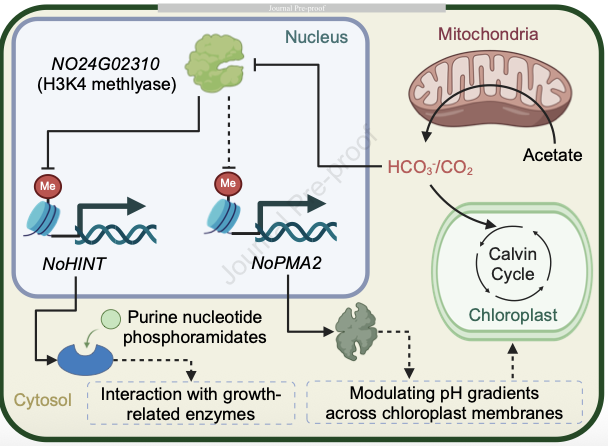
The study focused on Nannochloropsis oceanica, tracking its epigenomic dynamics as it transitioned from an environment with 5% CO2 to one with just 0.01% CO2. Using multi-dimensional epigenomic sequencing techniques, the researchers discovered that global DNA methylation in the alga remained stable at 0.13%, effectively ruling out DNA methylation as a major driver of its low-CO2response. By contrast, H3K4me2 methylationwas found to be closely associated with 43.1% of the genes that respond to low-CO2 conditions. These genes include those critical to photosynthesis and ribosome biogenesis, two processes essential for the alga’s survival under carbon-limited stress. Further analysis revealed that H3K4me2 appears to regulate gene transcription by altering chromatin accessibility, a mechanism that aligns with its role as a central regulator of low-CO2 adaptation.
To validate their findings, the team used CRISPR/Cas9 gene-editing technology. They knocked out NO24G02310—a gene that encodes an H3K4 methyltransferase, the enzyme responsible for adding methyl groups to H3K4. The modified algae showed a roughly 22% reduction in growth rate and a 15% decrease in biomass. Additionally, the levels of another histone modification (H3K4me1) dropped, and the genome-wide localization of H3K4me2 shifted—providing direct evidence of H3K4me2’s role in low-CO2 adaptation. Further experiments uncovered that H3K4 modification may act via two pathways: by regulating enzyme networks and by modulating chloroplast transmembrane pH gradients. Both mechanisms work to optimize the alga’s use of available CO2, enhancing its survival under low-carbon conditions.