https://doi.org/10.1016/j.biortech.2025.133788
http://english.cas.cn/newsroom/research_news/life/202601/t20260114_1145714.shtml
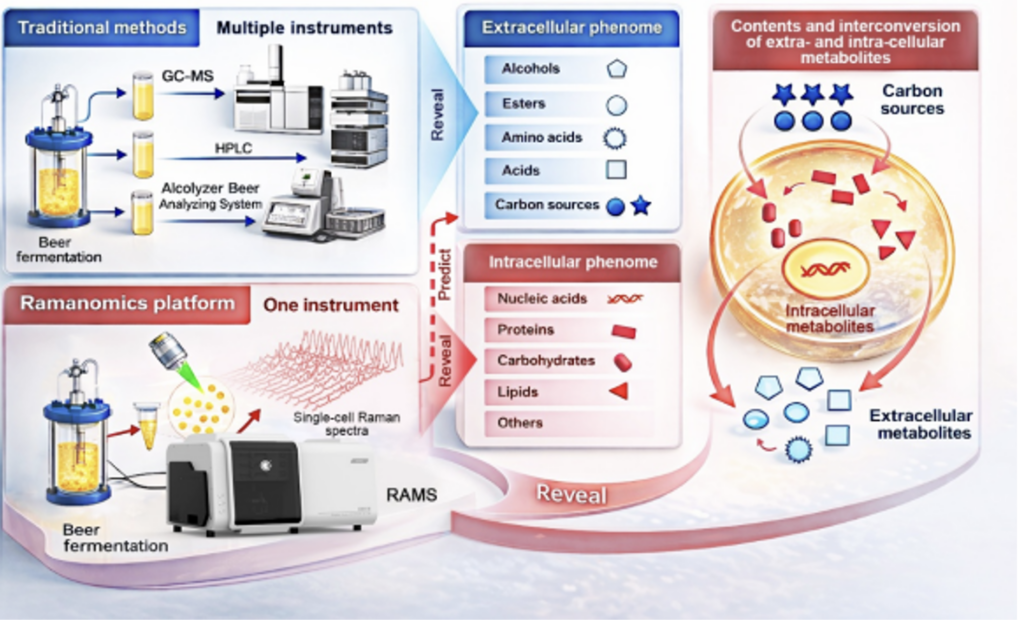
Breweries typically monitor fermentation by analyzing broth composition. Alcohols, esters, acids and residual sugars are quantified via chromatography-based assays. While reliable, these tests are time-consuming and only yield batch-average results.
A research led by scientists from the CAS Qingdao Institute of Bioenergy and Bioprocess Technology (QIBEBT) has simplified this process and developed a novel workflow dubbed “process ramanomics,” which is based on spontaneous single-cell Raman spectroscopy.
To validate the approach, the researchers tracked an industrial beer fermentation process using the lager yeast Saccharomyces pastorianus, sampling a single production batch over an eight-day period. At each stage of fermentation, they collected high-throughput Raman spectra from individual cells (a “ramanome”) and matched these unique molecular fingerprints to conventional lab measurements of 43 extracellular phenotypes in the fermentation medium.
Using multivariate regression analysis, the team found that ramanomes could accurately predict 19 extracellular phenotypes. This included four higher alcohols, four esters, four amino acids, two organic acids, four mono- and disaccharide substrates, and the alcohol-to-ester ratio—a commonly used indicator tied to beer flavor balance. In practical terms, a single, rapid cellular analysis can now replace multiple time-intensive chemical assays—without sacrificing single-cell resolution details.
Because the models output cell-level predictions, the researchers also tracked phenotypic heterogeneity over time. Different metabolite classes displayed distinct heterogeneity trajectories, and for several phenotypes higher heterogeneity tended to accompany lower metabolite levels—suggesting that dispersion among cells may be a useful process-state indicator.