https://www.cell.com/the-innovation/fulltext/S2666-6758(25)00380-7
https://www.cas.cn/syky/202511/t20251110_5088010.shtml
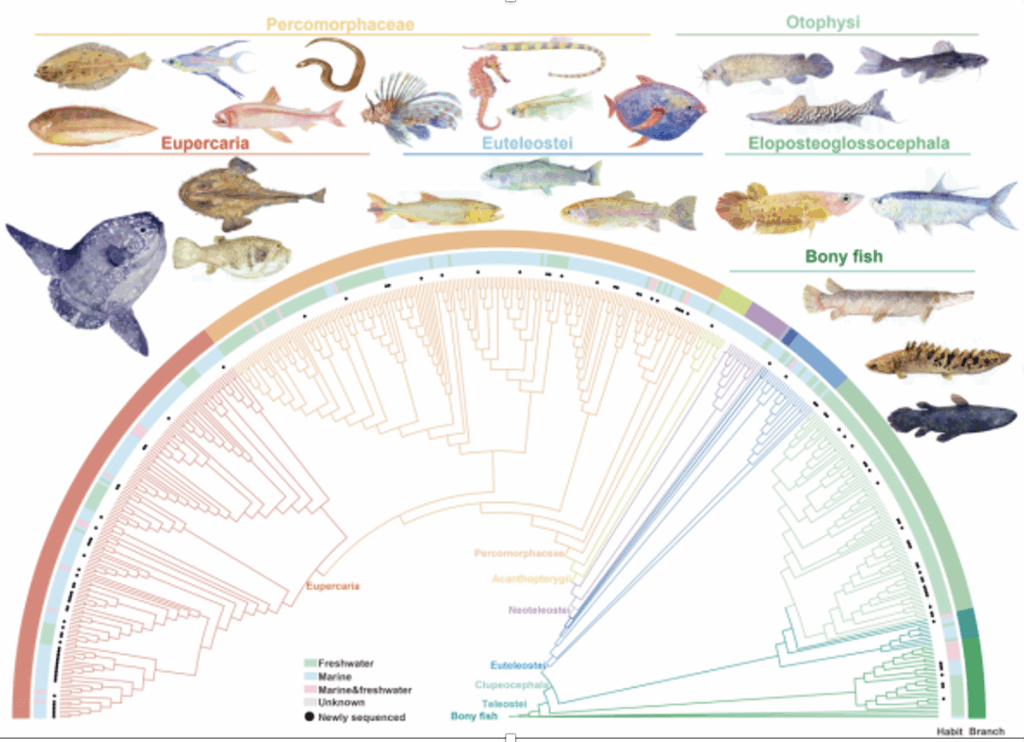
A team at the CAS Institute of Hydrobiology through de novo sequencing and integrated analysis of the whole genomes of 464 Eustemonia species, constructed the most comprehensive fish genome map to date, providing high resolution for elucidating the evolutionary history of fish.
The research team completed high-quality genome assembly for 110 new fish species, filled in three long-missing order-level taxonomic units for fish genome data, and integrated existing genomes to establish a whole-genome alignment matrix covering all 44 orders and a total of 464 species of teleost fish. This resource surpasses previous fish genomics studies in quantity and is comparable to large-scale mammalian and avian genome projects in terms of analytical precision, making it the largest and most comprehensive fish genome resource to date.
The study found that during the evolution of ray-finned fishes, the genome exhibited a gradual compression trend, particularly intron shortening, while exon length remained stable. The team identified nearly 30,000 highly conserved elements in teleost fishes, including 1,689 teleost-specific HCEs. These elements are associated with the development of key organs such as the brain, fins, heart, and gills, suggesting their regulatory role in morphological innovation in teleost fishes.
This study elucidated the genome structure, transposon dynamics, conserved and innovative elements, phylogenetic relationships, and evolutionary rates of teleost fishes, revealing the role of genome duplication and transposon activity in driving adaptive evolution in fish. These findings lay a data foundation for understanding the evolutionary patterns of vertebrate genomes, the origin of traits, and mechanisms of ecological adaptation.