https://www.cas.cn/cm/202411/t20241120_5039986.shtml
https://www.nature.com/articles/s42256-024-00920-9
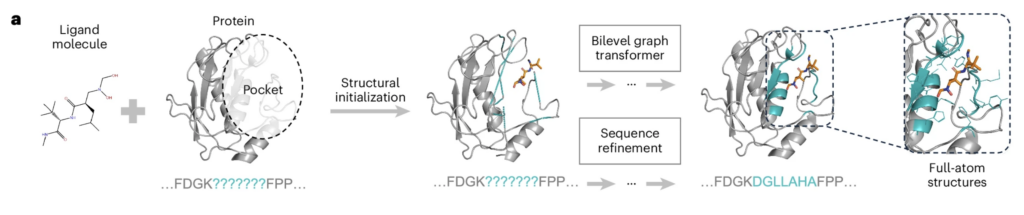
Professor Liu Qi of the University of Science and Technology of China and Professor Marinka Zitnik’s research group at Harvard University have collaborated to design a deep generation algorithm PocketGen to generate protein pocket sequences and spatial structures that bind to small molecules.
To design protein pockets, the traditional method is energy optimization and template matching, but this method has slow computational efficiency and low success rate. Existing deep learning-based models are not designed specifically for studying protein-small molecule interactions and may not be able to fully describe the characteristics between protein and small molecule binding pocket regions.
PocketGen provides a new solution to the above problems. The algorithm consists of two parts: a double-layer graph Transformer encoder and a protein pre-trained language model. The two parts correspond to the structural information and sequence information of the protein, respectively. Through the simultaneous information processing and continuous iteration of the two parts, the algorithm finally generates the required protein pocket.
The authors claim that PocketGen has outstanding performance in terms of computational efficiency and the success rate of protein pocket design.