https://www.cas.cn/syky/202411/t20241111_5039063.shtml
https://www.sciencedirect.com/science/article/abs/pii/S0167779924002816
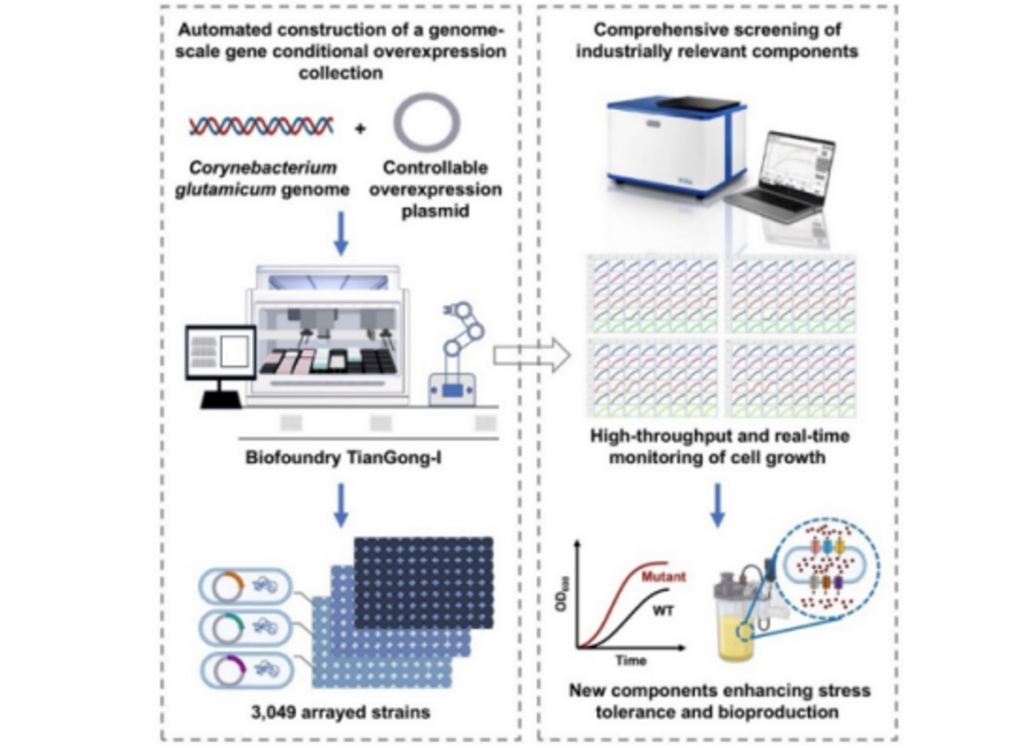
Corynebacterium glutamicum is the main bacterium in the industrial production of amino acids. However, the functions of about 50% of the genes in the genome of Corynebacterium glutamicum are still unclear.
The CAS Tianjin Institute of Industrial Biotechnology has achieved the automated construction and high-throughput screening of a single gene overexpression library on a whole genome scale in Corynebacterium glutamicum, and identified multiple new functional elements that affect osmotic pressure tolerance and amino acid production. This study used the Tianjin Institute of Industrial Biotechnology Biofoundry to construct a single gene overexpression library covering 99.7% of the whole genome of Corynebacterium glutamicum, and established a resource sharing website. At the same time, the study established a high-throughput screening method for new functional elements through fully automatic and real-time growth curve determination; 15 new elements that can improve osmotic pressure tolerance and L-lysine production, such as new transcriptional regulatory factors and DNA repair proteins, were obtained through full-library screening, and new mechanisms of osmotic pressure tolerance were revealed. Further, the study screened nearly 400 membrane transporter sub-libraries and obtained efficient and specific new L-threonine efflux proteins in Corynebacterium glutamicum and Escherichia coli. The study applied the new efflux protein to the transformation of L-threonine production strains, achieving the highest level of L-threonine production in Corynebacterium glutamicum.
As one of the application scenarios of the bio-foundry, the study provides a systematic strategy for the screening of new functional elements. This strategy can be extended to the creation and research of other industrial microorganisms and high-level industrial strains. At the same time, the whole genome-scale strain library constructed by this study is expected to become an industrial strain resource for basic biological research and amino acid development.