https://www.cell.com/cell/abstract/S0092-8674(24)00469-0
https://www.cas.cn/syky/202405/t20240526_5015780.shtml
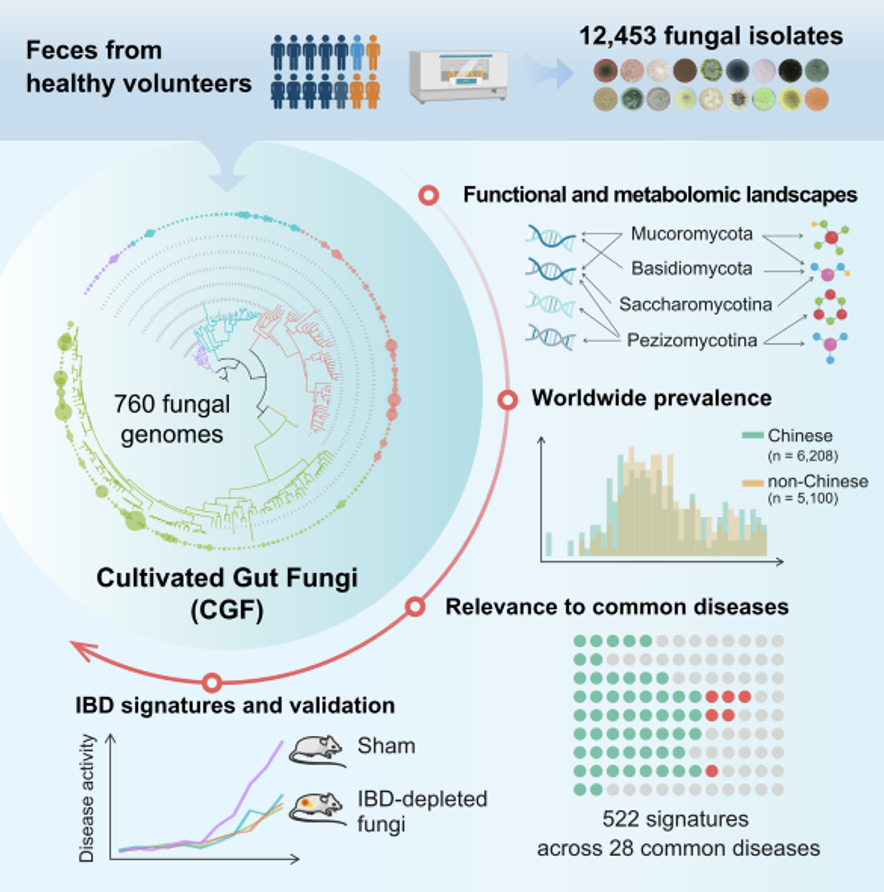
A Chinese-French team has published a genomic compendium of cultivated human gut fungi that characterizes the gut mycobiome and its relevance to common diseases. The human digestive system is composed of a variety of microorganisms, including bacteria, archaea, fungi and viruses. At present, there have been many studies on the community structure and biological functions of human intestinal bacteria. As an important part of the intestinal microecology, the intestinal fungal community affects various physiological and pathological processes of the host. This study used culture group technology to culture 12,453 fungal strains from fecal samples of 135 healthy Chinese people and performed whole-genome sequencing. 760 intestinal fungal genomes (covering 48 families and 206 species) were obtained, 69 of which were identified for the first time. The study conducted functional gene analysis and targeted metabolomics determination on 206 species of fungi, and elaborated on the characteristics of intestinal fungi in terms of gene functions and metabolites. Furthermore, the study analyzed 11,000 human fecal metagenomic data and revealed the structural characteristics of intestinal fungal communities in Chinese and non-Chinese populations. The study analyzed the characteristics of the intestinal fungi group of 28 diseases, identified disease-related intestinal fungal signature signals, and focused on the intestinal fungal characteristics of patients with inflammatory bowel disease. The characteristics of the target strains were verified in various mouse colitis models. Biological functions, some active substances and molecular mechanisms.