https://www.nature.com/articles/s41589-023-01489-x
https://english.cas.cn/newsroom/research_news/life/202307/t20230731_334150.shtml
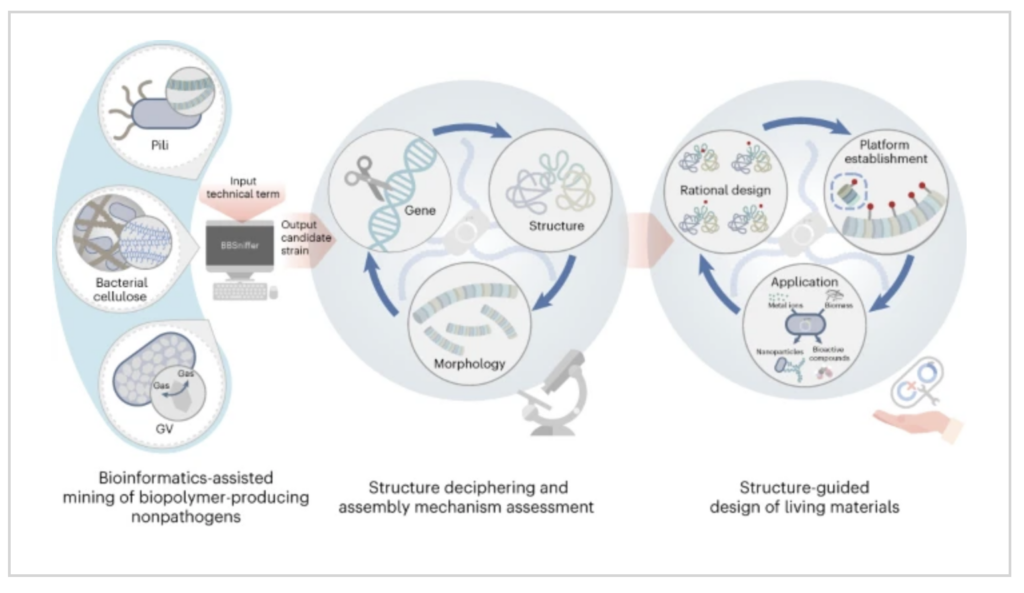
A research team from the Shenzhen Institute of Advanced Technology (SIAT) of the Chinese Academy of Sciences has developed an integrated technological workflow for engineered living materials ELMs by synergistically combining tools from bioinformatics, structural biology, and synthetic biology. The workflow enables the exploration, comprehension, and utilization of bacterial biopolymers of interest and suitable biopolymer-producing bacteria as structural building blocks, and then hosts for systematic and sequential design of living materials.
They developed a technological workflow for facilitating ELMs design by rationally integrating bioinformatics, structural biology and synthetic biology technologies. With a bioinformatics software, termed Bacteria Biopolymer Sniffer (BBSniffer), they succeeded in the fast mining of biopolymers and biopolymer-producing bacteria of interest. As a proof-of-principle study, using existing pathogenic pilus as input, they identified the covalently linked pili (CLP) biosynthetic gene cluster in the industrial workhorse Corynebacterium glutamicum. Genetic manipulation and structural characterization revealed the molecular mechanism of the CLP assembly, ultimately enabling a type of programmable pili for ELM design. Finally, engineering of the CLP-enabled living materials allowed them to transform cellulosic biomass into lycopene by coupling the extracellular and intracellular bioconversion ability.