https://www.cas.cn/syky/202308/t20230831_4967205.shtml
https://www.science.org/doi/10.1126/science.abq7487
Changes in prehistoric population size reflect changes in climatic environments during that period, and thus retrospectively through population genetics research methods can provide a deeper understanding of the formation of modern humans. The available fossil record suggests that the last one million years was a critical period of human evolution, but the study of human population history is mostly confined to the last 300,000 to 100,000 years. Although ancient DNA sequencing technology has developed rapidly in recent years, it is not possible to extract ancient DNA from 300,000-year-old African human ancestor fossils due to hot conditions that are not conducive to DNA preservation.Therefore, this study analyzes the genomes of modern populations through the new theory of population genetics and explores the history of populations that existed millions of years ago.
Although there was no written record of group size in prehistory, effective group size affects the rate of ancestry tracing in each generation, that is, the probability that two lineages came from the same ancestor in the previous generation. Thus, human ancestors had left imprints in the group genome reflecting the group size at that time. The older the group, the weaker the imprinting signals that remain to this day. In order to accurately interpret these signals, and thus accurately estimate human population history millions of years ago, researchers have created a new theory of population genetics and computational biology, the Fast Very Small-Time Ancestry FitCoal. Following the mathematical derivation of this theory, this work obtains an analytical solution for the expected value of the length of the ancestry dendrites corresponding to each mutation type (i.e., the mutation frequency spectrum) under an arbitrary population model, as well as the exact likelihood of the probability of observing a sample mutation spectrum under population history conditions. As a result, FitCoal can quickly and automatically search for the maximum likelihood value without prior knowledge of the group history, thus estimating the group history and conducting a “census” of ancient human populations.
The accuracy of an analytical method can be judged by setting up a true model and analyzing the DNA polymorphism data generated by the simulation, measuring the unbiasedness and 95% confidence interval of the estimated group history. The population history estimated by FitCoal is not only unbiased, but also has smaller confidence intervals than the PSMC, Stairway Plot and SMC++ methods that are commonly used in the field today. The study further analyzes population histories under different conditions, including population mingling and natural selection, through extensive computer simulations. The results all show that FitCoal can accurately estimate human group history over millions of years.
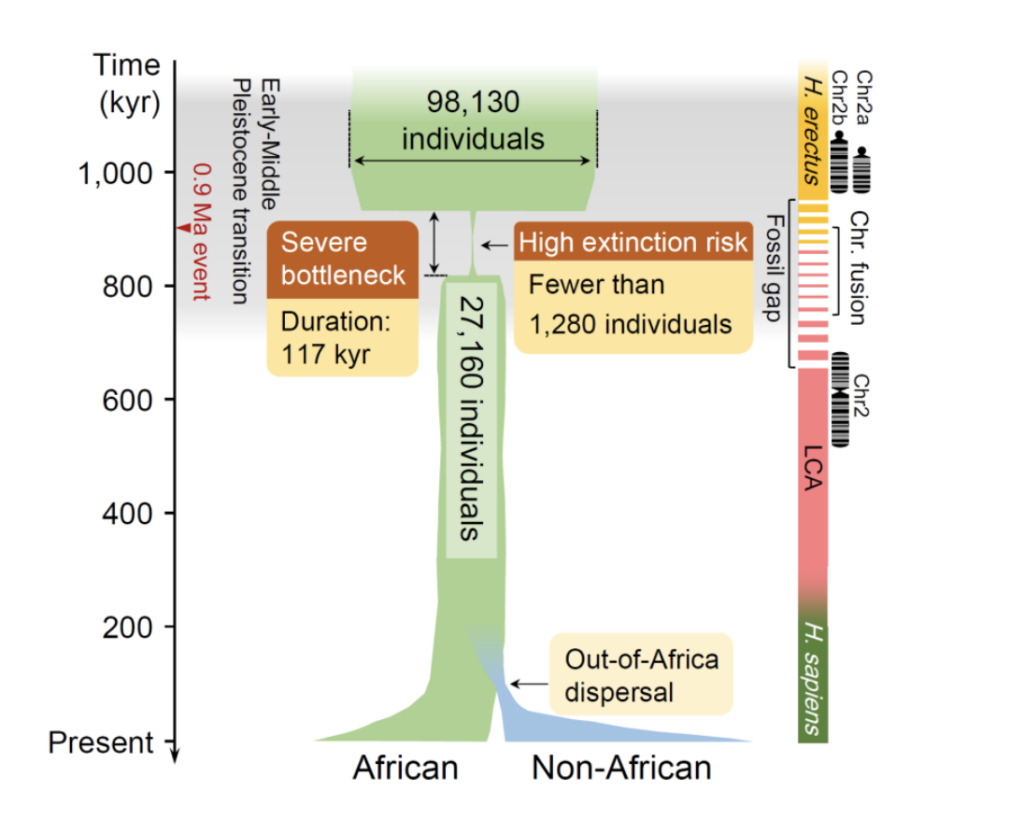
Based on FitCoal, the study further analyzed genomic data from a total of 50 modern human populations generated by the Thousand Genomes Project and the HGDP-CEPH Genome Project, and found for the first time that 930,000 years ago, the human ancestor almost became extinct due to the dramatic climatic changes in the Early and Middle Pleistocene transitions that resulted in the loss of about 98.7% of its individual members in a short period of time. The average number of adult individuals over a period of 117,000 years was only 1,280, and two independent data sets from the Thousand Genomes and HGDP-CEPH yielded nearly identical valuations of this group size, 1,270 and 1,300, respectively.This average valuation represents the upper limit of the minimum group size during the ancient group bottleneck, taking into account natural fluctuations in group size. The study was further validated using two southern African groups from the HGDP-CEPH dataset. Although the sample sizes were only six and eight individuals, FitCoal still detected the teleost population bottleneck. The results of the resampling of the African populations show that FitCoal can detect this ancient population bottleneck with the genomes of only three African individuals, further demonstrating that computational biology innovations have made this discovery possible.
This severe ancient population bottleneck coincides with the missing link in the fossilized African human ancestor, the disappearance of the Homo erectus fossil, the formation of a new lineage of Archaeopteryx species (LCA), and the fusion stage of the two Archaeopteryx chromosome 2s. Further, this suggests that severe group bottlenecks during the Early and Middle Pleistocene transition had a critical impact on human evolution and may have determined the formation of many key phenotypes of modern humans. At the same time, the attenuation of population size during this ancient period reduced the genetic diversity of 65.85% of modern populations, with profound impacts on human life and health.
The study was led by the Institute of Nutrition and Health and East China Normal University, in collaboration with the University of Rome, Italy, the University of Florence, Italy, and the University of Texas, USA.